|
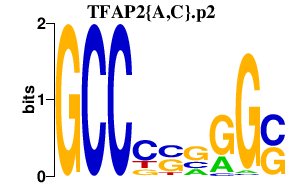
chr15_+_39573363
|
6.757
|
NM_002220
|
ITPKA
|
inositol 1,4,5-trisphosphate 3-kinase A
|
|
chr3_-_130807865
|
5.874
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr15_+_64782468
|
4.904
|
|
SMAD6
|
SMAD family member 6
|
|
chr19_-_11452439
|
4.841
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr15_-_81113676
|
4.830
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr4_+_2031036
|
4.808
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr12_+_109956210
|
4.480
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr3_-_129689394
|
4.334
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr19_+_540849
|
4.229
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr16_-_87535106
|
4.224
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr9_+_125813849
|
4.213
|
|
LHX2
|
LIM homeobox 2
|
|
chr9_+_94987032
|
4.129
|
NM_006648
|
WNK2
|
WNK lysine deficient protein kinase 2
|
|
chr1_+_1557419
|
3.941
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr12_+_2032649
|
3.846
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr5_+_170668892
|
3.751
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr2_+_120820140
|
3.711
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr8_-_144583718
|
3.582
|
NM_201589
|
MAFA
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian)
|
|
chr2_+_42128521
|
3.485
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr1_-_11674264
|
3.484
|
NM_001127325
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast)
|
|
chrX_+_68641802
|
3.422
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr10_-_88116181
|
3.404
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr8_+_104582151
|
3.354
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr3_+_182912769
|
3.254
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr10_+_94439658
|
3.225
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr17_+_62391216
|
3.209
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr4_-_41848874
|
3.198
|
|
BEND4
|
BEN domain containing 4
|
|
chr22_-_37569679
|
3.186
|
|
|
|
|
chr10_-_28074752
|
3.170
|
NM_173576
|
MKX
|
mohawk homeobox
|
|
chr5_+_149526533
|
3.157
|
NM_001804
|
CDX1
|
caudal type homeobox 1
|
|
chr14_-_64416307
|
3.128
|
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr1_+_41022066
|
3.113
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr17_+_56831951
|
3.032
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr2_-_47650973
|
3.020
|
NM_022055
|
KCNK12
|
potassium channel, subfamily K, member 12
|
|
chr18_+_53253775
|
3.020
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr3_-_124650081
|
3.006
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr12_+_52665196
|
2.989
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr10_-_131652364
|
2.989
|
|
EBF3
|
early B-cell factor 3
|
|
chr3_+_50217692
|
2.983
|
NM_006841
|
SLC38A3
|
solute carrier family 38, member 3
|
|
chr7_-_100331417
|
2.977
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr4_-_85638410
|
2.963
|
NM_006168
|
NKX6-1
|
NK6 homeobox 1
|
|
chr10_-_134449467
|
2.851
|
NM_177400
|
NKX6-2
|
NK6 homeobox 2
|
|
chr2_+_74595073
|
2.820
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr20_-_538909
|
2.810
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr17_+_45993338
|
2.780
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr22_-_37569962
|
2.775
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr10_+_23521464
|
2.761
|
NM_178161
|
PTF1A
|
pancreas specific transcription factor, 1a
|
|
chr9_-_93751900
|
2.758
|
NM_004560
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2
|
|
chr11_-_60819110
|
2.742
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr3_+_148610516
|
2.736
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr1_+_95355415
|
2.716
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr2_-_45090025
|
2.662
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr2_+_102602579
|
2.612
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr17_+_56832255
|
2.592
|
|
TBX2
|
T-box 2
|
|
chr20_-_21326046
|
2.585
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr20_+_20296744
|
2.579
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr4_+_4912272
|
2.561
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr16_+_2461500
|
2.553
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr17_-_77512346
|
2.534
|
NM_178493
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila)
|
|
chr19_-_14177980
|
2.517
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr10_-_103525646
|
2.517
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr17_-_10042579
|
2.495
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr5_-_11956964
|
2.492
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr17_+_29931880
|
2.473
|
NM_207313
|
TMEM132E
|
transmembrane protein 132E
|
|
chr8_+_1937711
|
2.471
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr2_-_42573980
|
2.446
|
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr3_+_71885890
|
2.443
|
NM_018971
|
GPR27
|
G protein-coupled receptor 27
|
|
chr20_+_19141364
|
2.441
|
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr19_+_12805747
|
2.437
|
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr1_+_22909916
|
2.422
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr9_+_128416542
|
2.417
|
NM_001174146
NM_001174147
NM_002316
|
LMX1B
|
LIM homeobox transcription factor 1, beta
|
|
chr1_-_31154023
|
2.388
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr17_-_44058612
|
2.382
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr1_-_50661716
|
2.380
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr5_-_11957127
|
2.359
|
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr7_-_150306169
|
2.345
|
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr3_+_129690735
|
2.307
|
|
|
|
|
chr13_+_24844084
|
2.306
|
NM_016529
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr22_+_28446288
|
2.293
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr15_+_39008822
|
2.291
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr1_-_6162669
|
2.287
|
NM_015557
|
CHD5
|
chromodomain helicase DNA binding protein 5
|
|
chr21_+_33364420
|
2.268
|
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr1_-_110414756
|
2.266
|
NM_006492
|
ALX3
|
ALX homeobox 3
|
|
chr4_-_174687104
|
2.245
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr16_+_85158357
|
2.243
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr1_+_11674365
|
2.243
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr16_+_88169612
|
2.238
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr9_-_119217147
|
2.231
|
|
ASTN2
|
astrotactin 2
|
|
chr2_+_171280944
|
2.229
|
|
SP5
|
Sp5 transcription factor
|
|
chr17_+_8865547
|
2.224
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr18_-_42590989
|
2.202
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr2_+_176680251
|
2.184
|
NM_021192
|
HOXD11
|
homeobox D11
|
|
chr22_+_43476710
|
2.180
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr11_-_60819273
|
2.178
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr2_+_176695658
|
2.165
|
NM_014213
|
HOXD9
|
homeobox D9
|
|
chr1_+_2975590
|
2.154
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr4_+_48180033
|
2.142
|
NM_152679
|
SLC10A4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4
|
|
chr1_+_235272324
|
2.129
|
NM_001035
|
RYR2
|
ryanodine receptor 2 (cardiac)
|
|
chr3_-_71886203
|
2.124
|
NM_001134649
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr15_+_30797466
|
2.116
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr20_-_62151216
|
2.112
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr13_+_111769913
|
2.111
|
NM_005986
|
SOX1
|
SRY (sex determining region Y)-box 1
|
|
chr22_+_43476755
|
2.101
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr9_+_125813672
|
2.100
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr2_-_236741352
|
2.092
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr16_+_166678
|
2.088
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr6_+_19945578
|
2.074
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr1_-_99242765
|
2.055
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr11_+_43920385
|
2.053
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr16_-_49742651
|
2.047
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr17_+_40655060
|
2.040
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr19_-_54557450
|
2.015
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr19_+_10392128
|
2.013
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr14_+_68796621
|
2.006
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr16_-_19803606
|
2.004
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr14_+_69021149
|
2.003
|
NM_001161498
|
UPF0639
|
UPF0639 protein
|
|
chr11_-_1549719
|
1.996
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr16_-_51138306
|
1.984
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr22_-_17891777
|
1.984
|
|
CLDN5
|
claudin 5
|
|
chr18_-_4445265
|
1.976
|
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr4_+_185064799
|
1.966
|
|
STOX2
|
storkhead box 2
|
|
chr11_-_69299351
|
1.957
|
NM_002007
|
FGF4
|
fibroblast growth factor 4
|
|
chr5_-_134397740
|
1.948
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr17_-_16335809
|
1.947
|
|
C17orf76
|
chromosome 17 open reading frame 76
|
|
chrX_-_139414890
|
1.945
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr13_-_101366995
|
1.940
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chr19_+_51059357
|
1.931
|
NM_004497
|
FOXA3
|
forkhead box A3
|
|
chr1_+_229365296
|
1.922
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr12_+_108636569
|
1.918
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr10_-_131652007
|
1.906
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr5_-_1348103
|
1.905
|
NM_001193376
NM_198253
|
TERT
|
telomerase reverse transcriptase
|
|
chr4_-_5945596
|
1.894
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr3_+_182912405
|
1.894
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr7_+_94374862
|
1.892
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr22_-_45311730
|
1.886
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr10_-_99780336
|
1.875
|
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr8_-_57521702
|
1.875
|
NM_001135690
|
PENK
|
proenkephalin
|
|
chr14_-_76677711
|
1.873
|
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chr6_-_110786167
|
1.851
|
NM_001123364
|
C6orf186
|
chromosome 6 open reading frame 186
|
|
chr7_-_123460722
|
1.837
|
NM_001136002
|
TMEM229A
|
transmembrane protein 229A
|
|
chr11_-_61441536
|
1.836
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr19_+_39664706
|
1.828
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr7_-_150305933
|
1.826
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr12_+_2032964
|
1.820
|
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr12_+_79995927
|
1.820
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr17_-_71747867
|
1.814
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr7_+_154943466
|
1.811
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr15_+_82113813
|
1.808
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr2_-_219633481
|
1.803
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr10_+_50488352
|
1.795
|
NM_003055
|
SLC18A3
|
solute carrier family 18 (vesicular acetylcholine), member 3
|
|
chr5_+_129268352
|
1.792
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr15_-_49174107
|
1.790
|
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr11_-_63292688
|
1.790
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr4_-_174686918
|
1.783
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr19_+_1704661
|
1.776
|
NM_001080488
|
ONECUT3
|
one cut homeobox 3
|
|
chr6_+_5030718
|
1.774
|
NM_001145115
|
PPP1R3G
|
protein phosphatase 1, regulatory (inhibitor) subunit 3G
|
|
chr20_-_61108831
|
1.765
|
NM_080606
|
BHLHE23
|
basic helix-loop-helix family, member e23
|
|
chr1_+_226937732
|
1.764
|
|
RHOU
|
ras homolog gene family, member U
|
|
chr9_-_95757365
|
1.764
|
NM_021570
|
BARX1
|
BARX homeobox 1
|
|
chr11_+_65311104
|
1.760
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr22_-_37180958
|
1.759
|
NM_152868
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4
|
|
chr7_-_35260213
|
1.745
|
NM_001077653
NM_001166220
|
TBX20
|
T-box 20
|
|
chr4_-_13155135
|
1.743
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr5_+_76542461
|
1.741
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr3_+_185580554
|
1.733
|
NM_003741
|
CHRD
|
chordin
|
|
chr1_+_63561632
|
1.732
|
|
FOXD3
|
forkhead box D3
|
|
chr2_+_241586927
|
1.728
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr6_-_117193505
|
1.727
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr2_-_42574587
|
1.719
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr1_-_114497994
|
1.718
|
NM_205848
|
SYT6
|
synaptotagmin VI
|
|
chr11_-_18769703
|
1.712
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr6_+_99389318
|
1.703
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr14_-_100104014
|
1.694
|
NM_001159531
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr4_-_123092358
|
1.692
|
NM_001130698
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr17_+_56832492
|
1.685
|
|
TBX2
|
T-box 2
|
|
chr9_+_966963
|
1.681
|
NM_021240
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chr2_+_172658453
|
1.677
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr11_+_63815348
|
1.674
|
NM_033310
|
KCNK4
|
potassium channel, subfamily K, member 4
|
|
chr8_+_56177567
|
1.671
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr2_+_176702667
|
1.669
|
NM_019558
|
HOXD8
|
homeobox D8
|
|
chr14_+_89597860
|
1.667
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr3_+_50687675
|
1.666
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr22_-_43783449
|
1.665
|
|
PHF21B
|
PHD finger protein 21B
|
|
chr8_-_71146022
|
1.663
|
|
PRDM14
|
PR domain containing 14
|
|
chr7_-_27206249
|
1.662
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr16_+_162845
|
1.660
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr10_+_29006425
|
1.659
|
NM_012342
|
BAMBI
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis)
|
|
chr10_+_115793795
|
1.654
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr1_-_111019051
|
1.653
|
|
|
|
|
chr3_-_50515857
|
1.651
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr10_+_101079099
|
1.643
|
|
CNNM1
|
cyclin M1
|
|
chr13_+_87122778
|
1.643
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr14_+_28306633
|
1.641
|
|
FOXG1
|
forkhead box G1
|
|
chr18_+_46340400
|
1.633
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr11_-_6633427
|
1.633
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr21_+_36993860
|
1.629
|
NM_005069
NM_009586
|
SIM2
|
single-minded homolog 2 (Drosophila)
|
|
chr17_-_76064842
|
1.628
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr7_-_45927263
|
1.628
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr16_+_52877009
|
1.623
|
|
|
|
|
chr3_-_129690056
|
1.620
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr4_+_42094612
|
1.618
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|